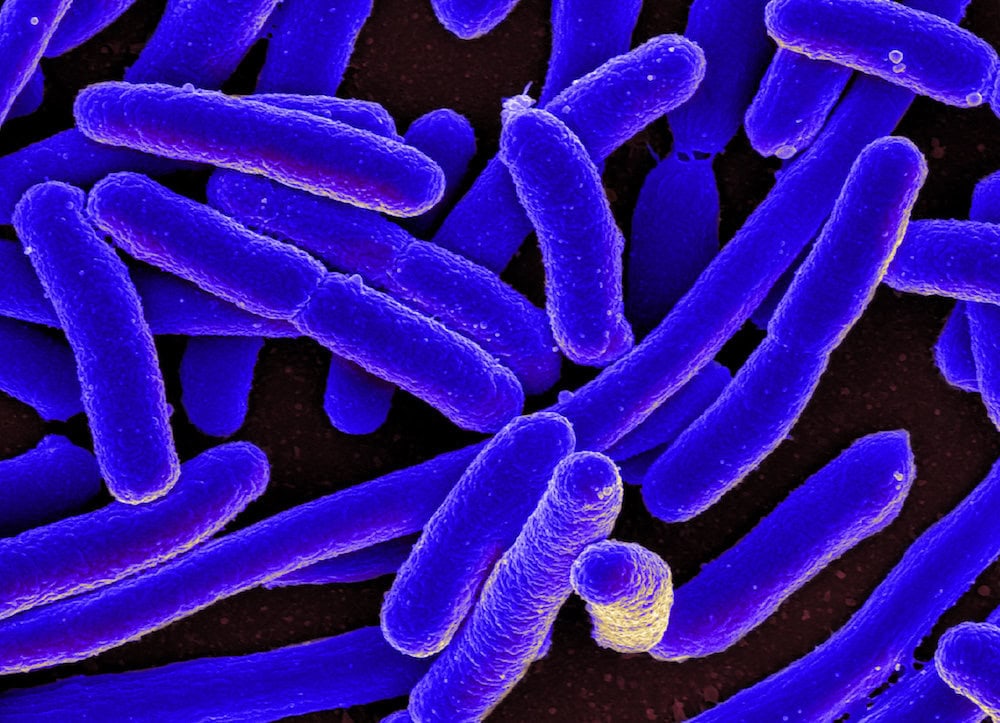
[Image above] Scanning electron micrograph of Escherichia coli, a type of bacteria that over the last 20 years has become increasingly drug resistant. Finding ways to destroy antibiotic resistant bacteria and antibiotic resistance genes is an important area of study for water treatment researchers. Credit: NIH Image Gallery, Flickr (CC BY-NC 2.0)
I have always taken it for granted that I can drink water straight from the tap. But in many places around the United States, such an assumption could lead to serious illness.
Lead contamination is one of the most well-known sources of contamination due to the water crisis in Flint, Michigan, a crisis that epitomizes the problem of unsafe tap water in the U.S. However, there are other equally dangerous contaminates threatening our water supply as well.
One example is per- and polyfluoroalkyl substances (PFAS), a group of more than 4,700 manmade chemicals that are notoriously difficult to break down. Last month, I talked about a unexpected discovery by Rice University researchers that found boron nitride serves as a surprisingly effective catalyst for degrading these chemicals.
Another contaminate of concern is antibiotic resistant bacteria, which have the ability to defeat drugs designed to kill them. Such bacteria have become an increasingly concerning problem around the world due in part to over-prescription and inappropriate prescription of antibiotics, practices which lead to bacteria developing resistance.
The agricultural sector is particularly notorious for its use of antibiotics. A 2014 report by the U.S. Food and Drug Administration revealed an estimated 80% of antibiotics sold in the U.S. are used in animals. These antibiotics are primarily meant to promote growth and prevent infection, but 70%–90% of antibiotics administered are excreted in urine and stool instead—and then widely dispersed into the environment through fertilizer, groundwater, and surface runoff.
Once in the environment, these antibiotics interact with bacteria and lead to growth of antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARGs), i.e., small pieces of DNA that carry genetic instructions for resistance from one germ to another. And often, these ARB and ARGs then end up in our water supply.
Water treatment systems are designed to filter out ARB and ARGs to ensure the water people drink is safe. However, though conventional systems such as chlorination are moderately effective in removing ARB, they are relatively ineffective at removing ARGs.
Thus, developing new treatment systems that effectively destroy ARGs is an active area of research. And that is a focus of George R. Brown Professor of Engineering Pedro J.J. Alvarez’s research group at Rice University.
Alvarez’s group is different from the group who conducted the boron nitride PFAS study mentioned above, but both groups are based in the Nanosystems Engineering Research Center for Nanotechnology Enabled Water Treatment (NEWT), a joint effort by Rice University, Arizona State University, University of Texas at El Paso, and Yale University to develop next-generation water treatment systems enabled by nanotechnology.
In recent papers published this March and July, Alvarez’s group focused on degrading ARGs through photocatalysis.
Photocatalysis: A brief explainer
Photocatalysis is the acceleration of a light-based chemical reaction in the presence of a catalyst. The acceleration is due mainly to the catalyst material, upon absorbing ultraviolet light, generating special oxygen-containing molecules called “reactive oxygen species” (ROS) that play a large role in photocatalytic degradation.
Photocatalysis has received significant attention lately as a potential eco-friendly disinfection process for eliminating waterborne microbial contaminants.
Molecular imprinting bolsters “trap-and-zap” degradation
The March paper looked to address a challenge that comes from using graphitic carbon nitride, an earth-abundant and metal-free photocatalyst, in photocatalysis processes to rid wastewater treatment plant effluent of ARGs.
“…degradation of ARGs by pristine or modified C3N4 can be adversely affected in [wastewater treatment plant] effluent, where soluble microbial product and natural organic matter compete with less abundant target contaminants (or ARGs in this case) for photogenerated ROS,” the researchers write.
In other words, reaction of graphitic carbon nitride with substances other than ARGs results in fewer ARGs being degraded.
If graphitic carbon nitride could be directed to react only with ARGs, then the rate of ARG degradation would improve. So the researchers looked to achieve this result by using a technique called molecular imprinting.
Molecular imprinting is known as a “lock and key” technique. A material is imprinted with template-shaped cavities (“locks”) to which only certain molecules (“keys”) can attach. Once the molecules attach, they are trapped in place and degraded through reaction with ROS.
The researchers found when they molecularly imprinted graphitic carbon nitride nanosheets, photocatalytic removal of the plasmid-encoded ARG blaNDM‑1 was 37 times faster than with bare graphitic carbon nitride.
“This trap-and-zap strategy significantly enhances removal of the eDNA [environmental DNA] gene, clearly outperforming commercial photocatalysts,” Alvarez says in a Rice press release.
The paper, published in Environmental Science & Technology, is “Selective adsorption and photocatalytic degradation of extracellular antibiotic resistance genes by molecularly-imprinted graphitic carbon nitride” (DOI: 10.1021/acs.est.9b06926).

(Left) Scanning electron micrograph showing the mesoporous structure of molecular-imprinted graphitic carbon nitride nanosheets. (Right) Transmission electron micrograph showing the sheet’s edge and its crystalline structure. Credit: Alvarez Research Group
Improved “wrap, trap, and zap” process for destroying ARGs
In the July paper, the researchers again used a “trap-and-zap” strategy to improve photocatalysis-induced degradation. But instead of using molecular imprinting to improve ARG adsorption, the researchers instead added a “wrap” step to the “trap-and-zap” strategy.
The researchers originally showed the potential of a “wrap, trap, and zap” strategy in 2014, when they wrapped nanosheet-assembled bismuth–oxygen–carbon microspheres in reduced graphene oxide to increase ROS production. (More ROS means more opportunities to degrade ARB and ARGs.)
“However, while enhanced ROS generation may improve disinfection, a higher rate of cells lysis [disintegration] increases the release of eARGs,” they write in the recent paper.
In other words, though more ROS were produced, ARB did not strongly attach to the microspheres. So, when the ARB degraded, the resulting ARGs spread into the environment rather than degrading as well.
To fix this problem, the researchers postulated that doping the reduced graphene oxide with nitrogen may improve ARB adhesion to the microspheres. And that is exactly what they found.
“The NRGO [nitrogen-doped reduced graphene oxide] shell increased the photocatalyst’s affinity toward the antibiotic resistant plasmid through π-π stacking and hydrogen-bond interfacial interactions, improving degradation of eARGs [environmental ARGs],” they write in the paper.
In addition, the NRGO shell served as a protective layer for the microspheres, “preventing photocorrosion under irradiation, thereby increasing the photocatalyst’s lifetime,” they add.
The paper, published in Water Research, is “Hierarchical Bi2O2CO3 wrapped with modified graphene oxide for adsorption-enhanced photocatalytic inactivation of antibiotic resistant bacteria and resistance genes” (DOI: 10.1016/j.watres.2020.116157).
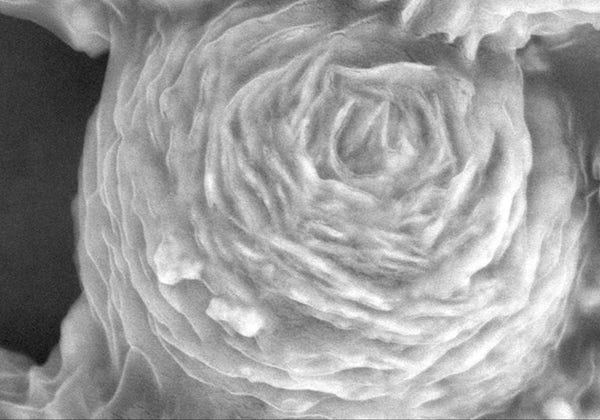
Scanning electron micrograph showing a nitrogen-doped reduced graphene oxide shell around layered bismuth–oxygen–carbon nanoplates. The microsphere traps and zaps antibiotic resistant bacteria and the resistance genes they release. Credit: Deyi Li, Tongji University
Author
Lisa McDonald
CTT Categories
- Environment
- Nanomaterials


