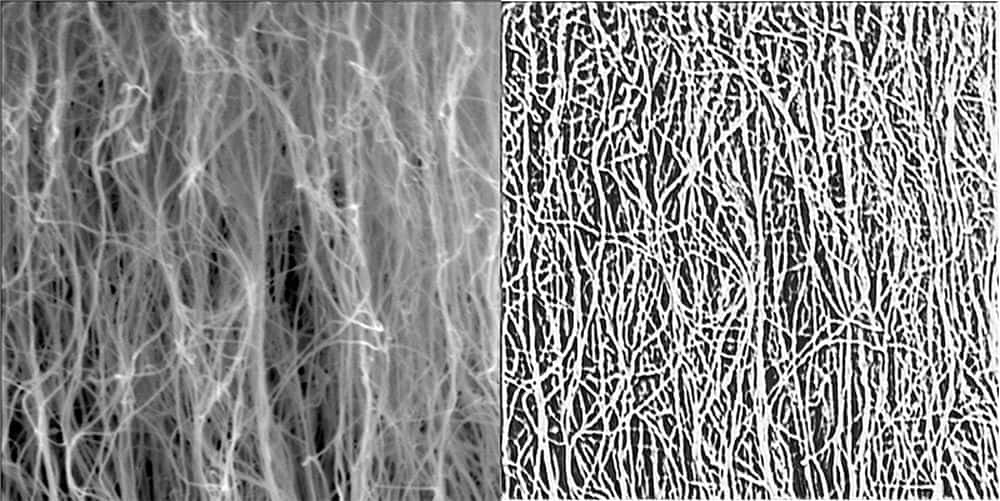
[Image above] Raw scanning electron microscopy image of a sample carbon nanotube forest and the output from the deep learning model. Credit: University of Missouri
Due to their tiny size, synthesizing isolated, individual carbon nanotubes (CNTs) is difficult and often impractical for device-level integration. So, CNTs are more frequently grown as “forests,” i.e., as collective arrays on a support substrate.
Though synthesizing numerous CNTs at once is easier than growing a single CNT, the tradeoff is that the CNT forest has vastly diminished properties due to the higher incidence of structural defects. For example, while the elastic modulus of an individual CNT is in the order of 1 TPa, the compressive elastic modulus of a CNT forest may be as low as 10 MPa.
Researchers looking to improve the physical properties of CNT forests can compare these traits to the forest’s growth parameters, which can guide modification of the synthesis process. Unfortunately, testing for physical properties of CNT forests often requires destruction of the forest, which prevents further data collection.
A method to determine physical properties of CNT forests indirectly using images would avoid the data limitation of current destructive testing methods. To develop such a method, the first step is creating a program capable of segmenting the dense forest into individual CNTs, which can then be analyzed for attributes such as orientation, linearity, density, and diameter.
Classical image processing approaches, such as thresholding and maximum entropy, used in previous studies only partially parsed out individual CNTs. Instead, emerging machine learning processes may support these methods and enable a better way to segment images for analysis.
In October 2022, University of Missouri (Mizzou) researchers presented their results at the European Conference on Computer Vision on using a deep learning technique to segment CNT forests in scanning electron microscopy images. The conference paper from this lecture published online in February 2023.
In the paper, they explain that because of data complexity and ambiguity, there is a shortage of high-quality datasets with associated labels that can enable use of supervised learning-based approaches.
So, “Self-supervising learning has emerged as an approach to learn good representations from unlabeled data and to perform fine-tuning with labeled features at the down-stream tasks,” they write.
As explained in a 2020 review paper, self-supervised learning models are trained using pseudo labels that are generated automatically without the requirement for human annotations. This training procedure consists of two steps: a pretext task and a downstream task. Feature representation is learned in the pretext task, while model adaptation and quality evaluation are completed in the downstream task.
The self-supervised segmentation method proposed by the Mizzou researchers consists of a novel deep neural network that uses two complementary sets of self-generated training labels.
- The first label, intensity thresholded raw input image, serves as a weak label that leads the network to perform binary CNT segmentation.
- The second label, CNT orientation histogram calculated directly from the raw input image, constraints the segmentation process by enforcing the network to preserve orientation characteristics of the original image.
These labels are used to conduct a two-component loss function.
- The first component is dice loss, which is computed between the predicted segmentation maps and the weak segmentation labels.
- The second component is mean squared error loss (MSE), which measures the difference between the orientation histogram of the predicted segmentation map and the original raw image.
The weighted sum of these two loss functions is used to train the deep neural network. Specifically,
- The dice loss forces the network to perform background–foreground segmentation using local intensity features.
- The MSE loss guides the network with global orientation features and leads to refined segmentation results.
After training the proposed network, the researchers compared its performance to three other segmentation methods: adaptive intensity thresholding, k-means clustering, and a recent unsupervised deep learning-based segmentation method based on differentiable feature clustering.
The researchers found their deep neural network results in more refined segmentation masks with better recall of the individual CNTs compared to the other three methods. Additionally, compared to the k-means clustering and unsupervised deep segmentation methods, the proposed network is more robust to illumination variations.
While this study focused on three CNT parameters—diameter, density, and growth rate—the researchers emphasize that the proposed network can be retrained using new datasets to improve performance or to adapt to new image characteristics. It could also be used to analyze other curvilinear structures, such as biological fibers or synthetic fibers.
In a Mizzou press release, coauthor Matt Maschmann, associate professor of mechanical and aerospace engineering and codirector of the MU Materials Science & Engineering Institute, says his dream for this research would be to develop an app or plug-in that allows anyone to scan a SEM image and see histograms representing the distributions of CNT diameters, growth rates, and even estimated properties.
“There’s a lot of work to do to make that happen, but it would be phenomenal. And I think it’s realistic,” he says.
The paper, published in Computer Vision–ECCV 2022 Workshops, is “Self-supervised orientation-guided deep network for segmentation of carbon nanotubes in SEM imagery” (DOI: 10.1007/978-3-031-25085-9_24).
Author
Lisa McDonald
CTT Categories
- Nanomaterials